Hello Iolite community!
I am writing to request your help with a problem I’m experiencing when importing data into Iolite 4.
We are using an Agilent 8900 ICP-MS running MassHunter 5.2 (version D01.02), coupled to a Cetac LSX-213 G2+ laser system.
During our experiment, a plasma shutdown occurred mid-run, so the sequence had to be completed over two days. When importing the data, we adjusted the timestamps in Iolite to align the files chronologically.
The issue is as follows:
The first three data files (from Day 1) import correctly as CSVs.
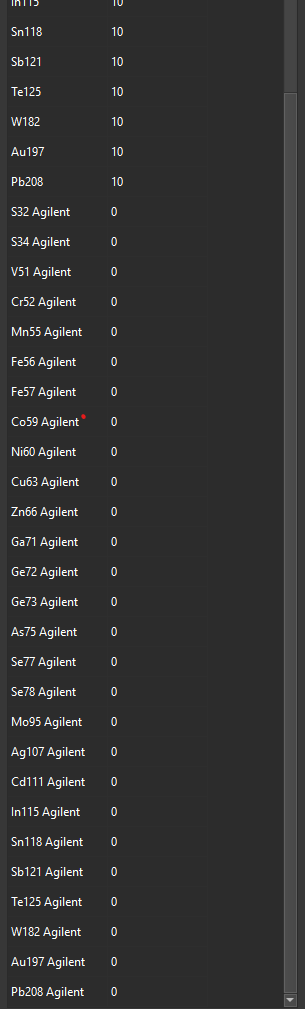
When we attempt to load the remaining files, Iolite creates additional isotopes labeled with “Agilent” suffixes (e.g., S32 Agilent), all with dwell times of 0.
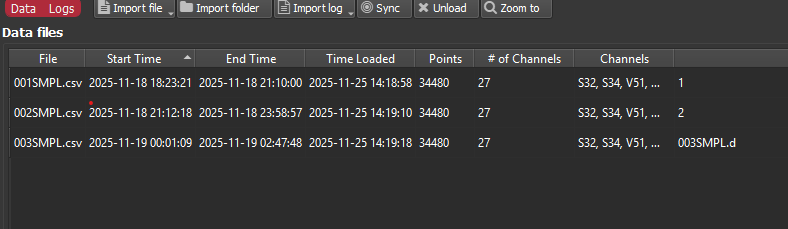
It also incorrectly identifies the CSV files that we want to upload as a “003SMPL.d, 004SMPL.dDear Iolite Support Team,
I am writing to request your help with a problem I’m experiencing when importing data into Iolite 4.
We are using an Agilent 8900 ICP-MS running MassHunter 5.2 (version D01.02), coupled to a Cetac LSX-213 G2+ laser system.
During our experiment, a plasma shutdown occurred mid-run, so the sequence had to be completed over two days. When importing the data, we adjusted the timestamps in Iolite to align the files chronologically.
The issue is as follows:
The first three data files (from Day 1) import correctly as CSVs.
When we attempt to load the remaining files (Day 2), Iolite creates additional isotopes labeled with “Agilent” suffixes (e.g., S32 Agilent), all with dwell times of 0.
It also incorrectly identifies the CSV files that we want to upload as a “003SMPL.d” and so on dataset, as if it were an Agilent MassHunter .d raw directory.
I have attached screenshots showing the issue.
Could you please advise what might be causing this issue and how we can resolve it?
Thank you in advance,
Estida Eensoo